Basic Tutorial Ib: Theorists¶
Theorists are classes designed to automate the construction of interpretable models from data. AutoRA theorists are implemented as sklearn regressors and can be used with the fit and predict methods.
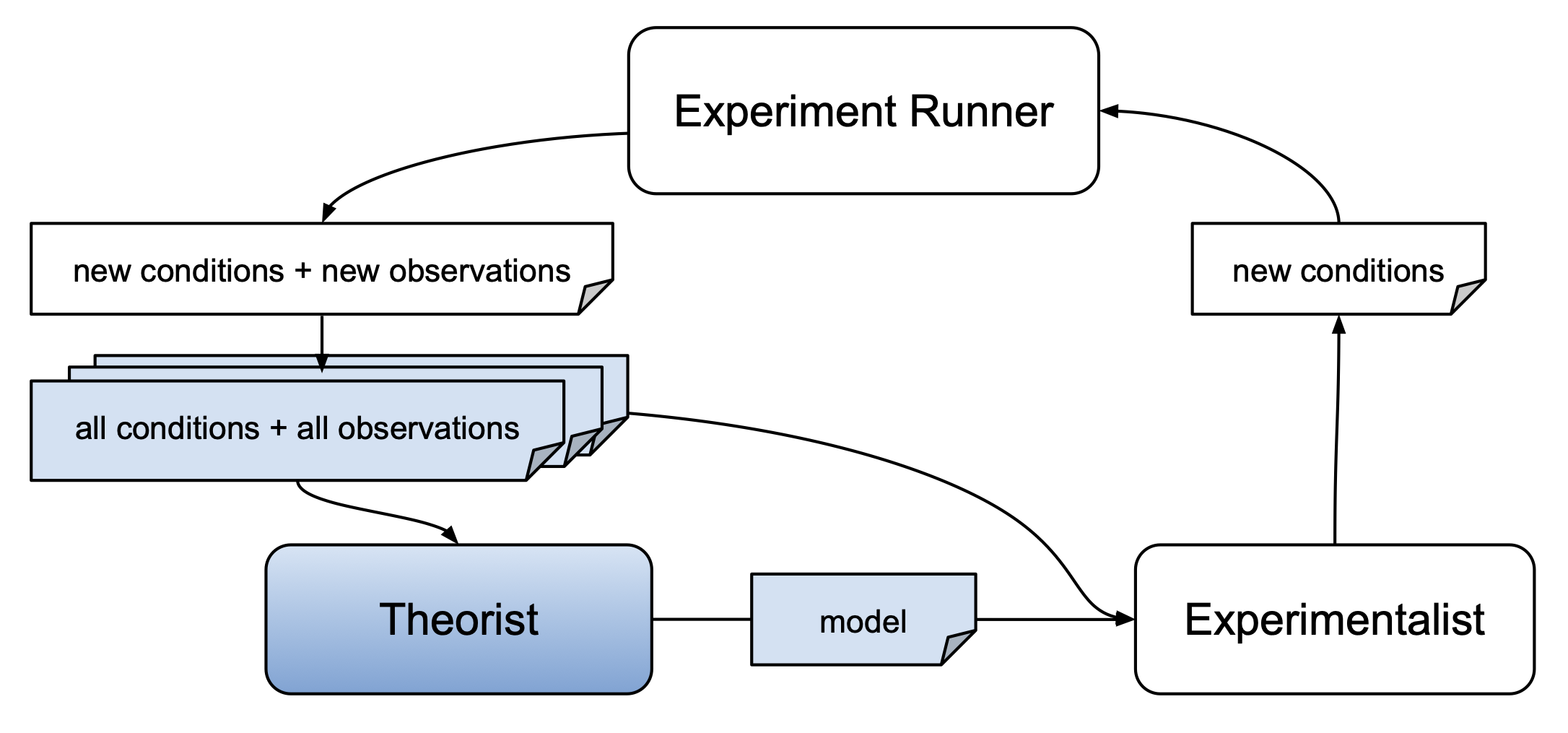
In order to use a theorist, you must first install the corresponding theorist package. Some theorists are installed by default when you install autora. Once a theorist is installed, you can instantiate it and use it as you would any other sklearn regressor. That is, you can call the fit function of the theorist by passing in experimental conditions and corresponding observations, and then call the predict function to generate predicted observations for novel experimental conditions using the discovered model.
The following tutorial demonstrates how to use the BMSRegressor (Guimerà et al., 2020, in Sci. Adv.)–a theorist that can discover an interpretable equation relating the independent variables of an experiment (experiment conditions) to predicted dependent variables (observations).
We will compare the performance of the BMSRegressor with two other methods: a polynomial regressor and a neural network regressor. The polynomial regressor is a simple model that can only fit polynomial functions, while the neural network regressor is a more complex model that can fit a wider range of functions. The BMSRegressor is a hybrid model that can fit a wide range of functions while also providing an interpretable, potentially non-linear equation.
Note: this tutorial requires Python 3.10 to run successfully.
Installing and Importing Relevant Packages¶
Install relevant subpackages from AutoRA
# fix default prior and release new package version
!pip install -q "autora[theorist-bms]"
Import relevant modules from AutoRA
from autora.theorist.bms import BMSRegressor
from autora.experiment_runner.synthetic.psychophysics.weber_fechner_law import weber_fechner_law
import numpy as np
from sklearn.base import BaseEstimator
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
Functions for Plotting Results¶
Before we begin, we also define some functions to plot the results of our models. Simply execute the following code block to define the plotting functions.
def present_results(model, x_test, y_test, arg='default', model_name=None, variable_names=None, select_indices=None, figsize=None, *args):
compare_results(models=[model], x_test=x_test, y_test=y_test, arg=arg, model_names=[model_name], variable_names=variable_names, select_indices=select_indices, figsize=figsize, *args)
def compare_results(models, x_test, y_test, arg='default', model_names=None, variable_names=None, observation_name=None, select_indices=None, figsize=None, *args):
if model_names is None or model_names == [None]:
names = ['Model '+str(i+1) for i in range(len(models))]
else:
names = model_names
if len(x_test.shape) == 1:
x_test = x_test.reshape(1, -1)
num_var = x_test.shape[1]
if variable_names is None:
var_names = ['Variable '+str(i+1) for i in range(num_var)]
else:
var_names = variable_names
if observation_name is None:
obs_label = 'Observations'
else:
obs_label = observation_name
match arg:
case 'default':
for i, model in enumerate(models):
print(model)
synthetic_runner.plotter(model)
case '2d':
if figsize is None:
size = (8,3)
else:
assert len(figsize) == 2 and isinstance(figsize, tuple), 'incorrect format for figure shape\nshould be tuple of form (i,j)'
size = figsize
for i, model in enumerate(models):
fig = plt.figure(figsize=size)
axes = []
y_predict = model.predict(x_test)
for j in range(num_var):
axes.append(fig.add_subplot(1, num_var, j+1))
axes[j].set_xlabel(var_names[j])
axes[j].set_ylabel(obs_label)
axes[j].set_title(names[i]+' fit on '+var_names[j])
axes[j].scatter(x_test[:,j], y_test, label='Ground Truth', alpha=0.5)
axes[j].scatter(x_test[:,j], y_predict, label='Predicted', alpha=0.5)
axes[j].legend()
for arg in args:
assert isinstance(arg, str), 'arguments must be in the form of a string'
try:
exec('axes[j].'+arg)
except:
raise RuntimeError(f'argument "{arg}" could not be executed')
fig.tight_layout()
plt.show()
case '3d':
if figsize is None:
size = (15,5)
else:
assert len(figsize) == 2 and isinstance(figsize, tuple), 'incorrect format for figure shape\nshould be tuple of form (i,j)'
size = figsize
axes = []
fig = plt.figure(figsize=size)
if select_indices is None:
idx = (0,1)
else:
len(select_indices) == 2 and isinstance(select_indices, tuple), 'incorrect format for select_indices\nshould be tuple of form (i,j)'
idx = select_indices
for i, model in enumerate(models):
y_predict = model.predict(x_test)
ax = fig.add_subplot(1, 3, i+1, projection='3d')
ax.w_xaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.w_yaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
ax.w_zaxis.set_pane_color((1.0, 1.0, 1.0, 0.0))
axes.append(ax)
axes[i].set_xlabel(var_names[idx[0]])
axes[i].set_ylabel(var_names[idx[1]])
axes[i].set_zlabel(obs_label)
axes[i].scatter(x_test[:, idx[0]], x_test[:, idx[1]], y_test, s=1, label='Ground Truth')
axes[i].scatter(x_test[:, idx[0]], x_test[:, idx[1]], y_predict, s=1, label='Predicted')
axes[i].set_title(names[i])
axes[i].legend()
axes[i].set_facecolor('white')
for arg in args:
assert isinstance(arg, str), 'arguments must be in the form of a string'
try:
exec('axes[j].'+arg)
except:
raise RuntimeError(f'argument "{arg}" could not be executed')
fig.tight_layout()
plt.show()
case 'choice':
for model in models:
y_pred = np.where(model.predict(x_test) > 0.5, 1, 0)
cm = confusion_matrix(y_true=y_test, y_pred=y_pred)
cmd = ConfusionMatrixDisplay(cm)
cmd.plot()
plt.show()
Our Study Object: The Weber-Fechner Law¶
We will evaluate our models to recover Weber-Fechner law. The Weber-Fechner law quantifies the minimum change in a stimulus required to be noticeable. Similar to Steven's power law, the greater the intensity of a stimulus, the larger the change needed to be perceivable. This relationship is hypothesized to be proportional to the logarithm of the ratio between the two stimuli:
$y = c \log\left(\dfrac{x_1}{x_2}\right)$
Generating Data From a Synthetic Psychophysics Experiment¶
Here, we leverage a synthetic experiment to generate data from this equation. It is parameterized by the constant $c$. The independent variables are $x_1$ and $x_2$, corresponding to the intensity of a stimulus and the baseline stimulus intensity, respectively. The dependent variable is $y$ perceived stimulus intensity.
constant = 3.0
# synthetic experiment from autora inventory
synthetic_runner = weber_fechner_law(constant=constant)
# experiment meta data:
synthetic_runner.variables
First, let's take a look at the variables in the synthetic experiment.
# independent variables
ivs = [iv.name for iv in synthetic_runner.variables.independent_variables]
# dependent variable
dvs = [dv.name for dv in synthetic_runner.variables.dependent_variables]
ivs, dvs
We can obtain the experimental data by running the synthetic experiment. The conditions contain values for the independent variables. Once we have the conditions, we can run the experiment to obtain the experiment_data containing both the conditions and the observations from the synthetic experiment.
# experimental data
conditions = synthetic_runner.domain()
experiment_data = synthetic_runner.run(conditions, added_noise=0.01)
# observations
observations = experiment_data[dvs]
experiment_data
Next, we split the data into training and testing datasets.
# split into train and test datasets
conditions_train, conditions_test, observations_train, observations_test = train_test_split(conditions, observations)
Fitting Models to the Data¶
In this section, we will fit the data with three techniques:
- Polynomial Regressor
- Neural Network Regressor
- Bayesian Machine Scientist
The last technique is an equation discovery algorithm implemented in the AutoRA framework.
We will repeat the following steps for each method:
- Initialize Model
- Fit Model to the Data
- Plot the Results
The polynomial regressor has a high interpretability, but it is limited in expressivity. It can only fit polynomial functions.
We first initialize the polynomial regressor. The polynomial regressor fits a polynomial function to the data. Below, we set the degree of the polynomial to 3.
Note that the PolynomialRegressor class is a simple implementation of a polynomial regressor using sklearn's PolynomialFeatures and LinearRegression classes.
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
class PolynomialRegressor:
"""
This theorist fits a polynomial function to the data.
"""
def __init__(self, degree: int = 3):
self.poly = PolynomialFeatures(degree=degree, include_bias=False)
self.model = LinearRegression()
def fit(self, x, y):
features = self.poly.fit_transform(x, y)
self.model.fit(features, y)
return self
def predict(self, x):
features = self.poly.fit_transform(x)
return self.model.predict(features)
Let's initialize the model
np.random.seed(0)
poly_model = PolynomialRegressor(degree=3)
And fit it to the training data, consisting of the experimental conditions and corresponding observations of our synthetic experiment.
poly_model.fit(conditions_train, observations_train)
Finally, we can plot the results of the polynomial regressor to evaluate the model's performance.
present_results(model=poly_model, x_test=conditions_test, y_test=observations_test)
present_results(model=poly_model, x_test=conditions_test, y_test=observations_test, arg='2d')
present_results(model=poly_model, x_test=conditions_test, y_test=observations_test, arg='3d')
In summary, we can see that the polynomial regressor is a simple model. It has high interpretability which allows us to quantify the law underlying the data in terms of a polynomial. However, it has some trouble fitting the data generated from the logarithmic psychophysics law due to its limited expressivity.
Neural networks are known for their high expressivity, allowing to fit any function. However, they are often considered black-box models due to their complex structure, limiting interpretability for the user.
For this section, we are using torch: an open-source machine learning library. It provides a flexible and dynamic computational graph, allowing for complex neural network architectures.
!pip install -q torch
We can now initialize a simple multi-layer perceptron (MLP) regressor using the MLPRegressor class from the sklearn.neural_network module. We will train it for a maximum of 500 iterations.
from sklearn.neural_network import MLPRegressor
import torch
nn_model = MLPRegressor(random_state=1, max_iter=500)
Similar to the polynomial regressor above, we can fit the neural network regressor to the training data.
np.random.seed(0)
torch.manual_seed(0)
nn_model.fit(conditions_train, observations_train)
And plot the results of the neural network regressor.
synthetic_runner.plotter(nn_model)
We may observe that the neural network regressor does a better job of fitting the data but it is less interpretable than the polynomial regressor. The neural network is a more complex model that can fit a wider range of functions, but it is also a black-box model that does not provide an interpretable equation, limiting its utility for scientific discovery.
Bayesian Machine Scientist¶
Expressivity: Medium
Interpretability: High
The Bayesian Machine Scientist (BMS) is one of the theorists that comes with the autora package. It is an equation discovery method that can fit a wide range of functions while providing an interpretable equation. It uses MCMC-Sampling to explore the space of possible equations and find the best-fitting equation for the data while minimizing the number of parameters in the equation.
We initialize the BMS regressor to run 1500 epochs.
bms_model = BMSRegressor(epochs=1500)
We can also fit the model to the training data...
bms_model.fit(conditions_train, observations_train, seed=0)
...and plot the results.
synthetic_runner.plotter(bms_model)
In addition, we can print the discovered equation.
print(bms_model)
Note that BMS is not only producing good fits but is also capable of recovering the underlying equation of the synthetic data, the Weber-Fechner law.
In summary, the BMS regressor is a powerful tool for fitting a wide range of functions while providing an interpretable equation. It strikes a balance between expressivity and interpretability, making it a valuable tool for scientific discovery. In our case, it is capable of re-discovering the Weber-Fechner law from the synthetic data.
Comparing the Models¶
Finally, we can compare all three fitted models in terms of their fit to the hold-out data using the compare_results function. This function plots the ground truth and predicted values for each model, allowing us to visually compare their performance.
models = [poly_model, nn_model, bms_model]
names =['poly_model', 'nn_model', 'bms_model']
compare_results(models=models, x_test=conditions_test, y_test=observations_test, model_names=names, arg='2d')
We can also compare the models in 3D space.
compare_results(models=models, x_test=conditions_test, y_test=observations_test, model_names=names, arg='3d')
Summary¶
In this tutorial, we compared the performance of three different theorists: a polynomial regressor, a neural network regressor, and the Bayesian Machine Scientist (BMS) regressor. The polynomial regressor is a simple model that can only fit polynomial functions, while the neural network regressor is a more complex model that can fit a wider range of functions. The BMS regressor is a hybrid model that can fit a wide range of functions while also providing an interpretable, potentially non-linear equation.
AutoRA provides interfaces for using theorists as sklearn regressors. This allows you to easily fit and evaluate theorists using the fit and predict methods. Note that such theorists may not be limited to fitting functions but may also discover complex computational models or algorithms describing the data.